
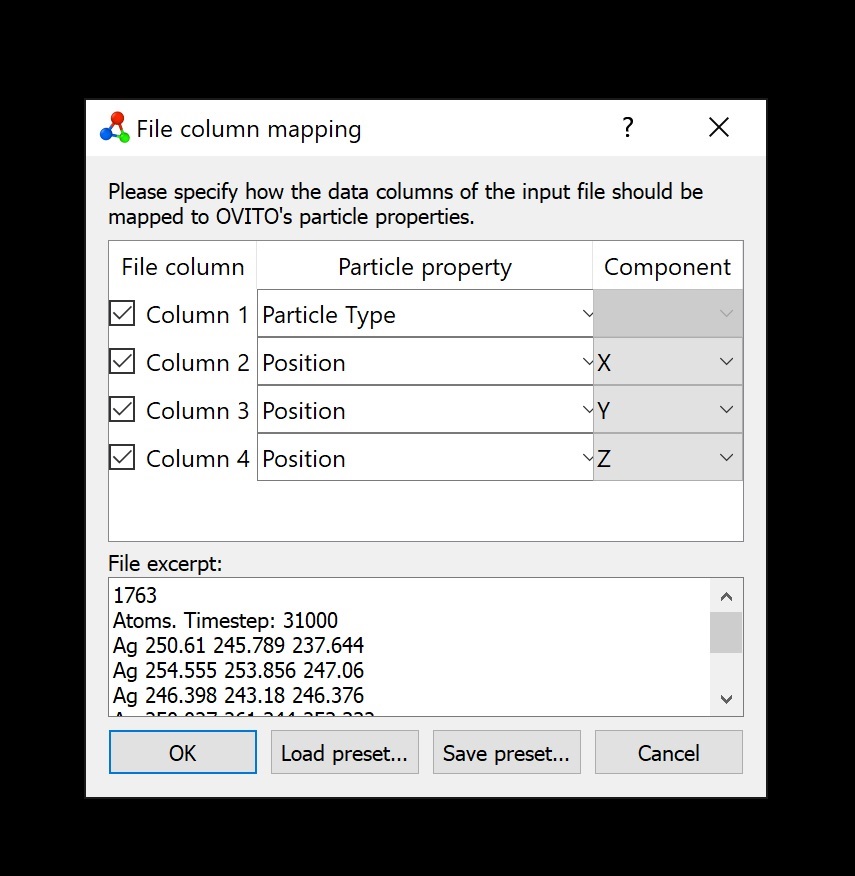
The XYZ reader in Jmol reads any of the following, alternative, formats in each atom's line (updated for Jmol v. Briefly, if the distance between two atoms is less than the sum of their covalent radii, they are considered bonded. Connectivity information is generated automatically for XYZ files as they are read into XMol-related applications. This intentional omission allows for greater flexibility: to create an XYZ file, you don't need to know where a molecule's bonds are you just need to know where its atoms are. Note that the XYZ format doesn't contain connectivity information. These components should be specified in angstroms. If an input line contains seven or eight fields, the last three fields are interpreted as the components of a vector. If an input line contains five or eight fields, the fifth field is interpreted as the atom's charge otherwise, a charge of zero is assumed. Optionally, extra fields may be used to specify a charge for the atom, and/or a vector associated with the atom. This line may be blank, or it may contain some information pertinent to that particular step, but it must exist, and it must be just one line long.Įach line of text describing a single atom must contain at least four fields of information, separated by whitespace: the atom's type (a short string of alphanumeric characters), and its x-, y-, and z-positions. The second line of the header leaves room for a descriptive string. Note, however, that if there is any text after the number Jmol will not automatically recognize it as an xyz-formatted file in that case, you need to explicitly indicate the format in the load command, like this: load xyz::myFile.xyz That given, the presence of any text after the number in the 1st line will not affect proper file reading. This integer may be preceded by whitespace anything on the line after the integer is ignored. The first line of a step's header is the number of atoms in that step. Each step is represented by a two-line "header," followed by one line for each atom.
The XYZ format supports multi-step datasets. Once data is in XYZ format, it may be examined by XMol, or converted to yet another format. In order to read the data into XMol, it would be possible to modify the datafile, perhaps by creating a shell script, so that it fit the relatively lenient requirements of the XYZ format specification. For example, suppose a molecular datafile was in a format not supported by XMol.

LOAD DIRECTORY OF XYZ IN JMOL SERIES
This simple, stripped-down, ASCII-readable format is intended to serve as a "transition" format for the XMol series of applications. XYZ datafiles specify molecular geometries using a Cartesian coordinate system.


 0 kommentar(er)
0 kommentar(er)
